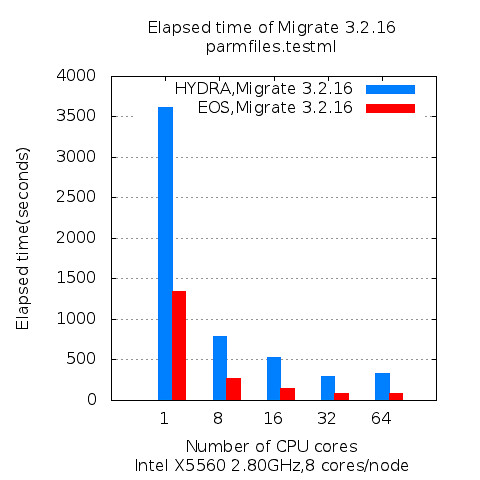
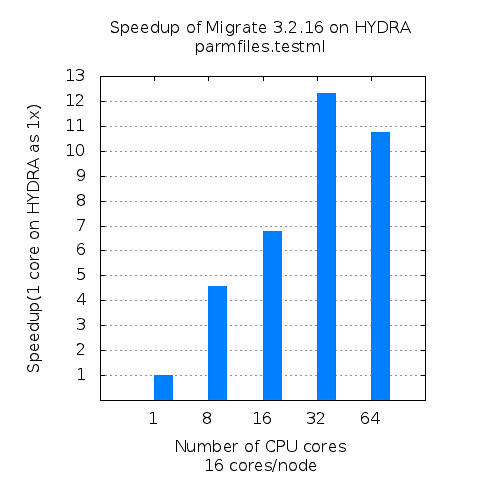
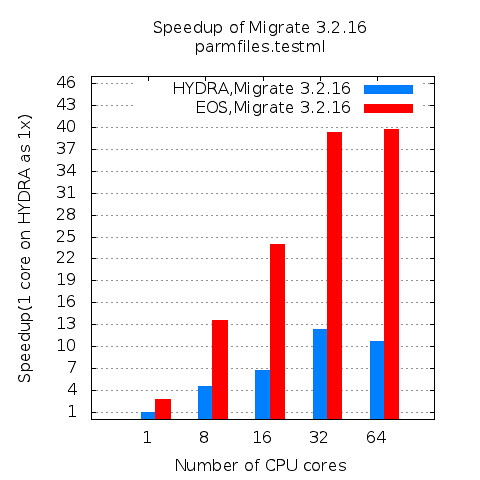
As GPU become readily available resources in high performance computing environment, more and more applications are being considered to be a target application for taking advantage of highly parallel computing capability of GPU. To evaluate the performance improvement in application level, molecular dynamics applications, NAMD, is experimented in this article and its performance has been analyzed in three different perspectives: Scalability, speedup, and GPU utilization.
1. Experimental environment
GPU nodes are relatively new addition to the existing EOS system at Texas A&M University. Table 1 shows detail of the node tested in this experiment. It has two six core 2.8Ghz Intel Xeon X5660 processors based on Westmere architecture and 24GB DDR3 DRAM with 1,333Ghz clock cycle. Additionally, it is equipped with two NVIDIA M2050 GPU devices. Each M2050 GPU device runs at 1.15Ghz and 2687MB GDDR5 memory with 1.546Ghz clock cycle. M2050 is a high performance grade device with 14 streaming multiprocessors and each of them has 32 cores which is based on Fermi architecture. It also has ECC memory error protection, and L1 and L2 cache to provide not only accuracy but also high performance on computation. The node is running 64-bits Red Hat Enterprise Linux Server release 5.4.
NAMD has been tested on three configurations. First, it is executed with CPU only configuration. Secondly, it is executed with 1 NVIDIA M2070 GPU. Finally, it is executed with 2 NVIDIA M2050 GPUs. Since these tests are submitted through batch system and there is no guarantee that each test ran exclusively on compute node and compute node could be shared by several other jobs as well. Therefore, the performance presented in this article could be different from exclusive testing environment.
|
|
NVIDIA
M2050
|
NVIDIA
M2070
|
INTEL
X5660
|
|
Clock
speed(GHz)
|
1.15
|
1.15
|
2.8
|
|
Memory(GB)
|
2.687
|
5.375
|
24.081
|
|
Memory
Clock(GHz)
|
1.546
|
1.566
|
1.333
|
|
# of CUDA
cores
|
448
|
448
|
N/A
|
|
CUDA Driver
ver.
|
4.0
|
4.0
|
N/A
|
|
CUDA CC ver.
|
2.0
|
2.0
|
N/A
|
To benchmark NAMD, apoa1 dataset is used in experiment. This dataset contains 92,000 atoms and simulates 500 steps. Among several parameters in input file, outputEnergies set to 100 as suggested in user manual to remove unnecessary CPU involvement for generating additional output in file.
2.Results
In this section, performance of NAMD has been analyzed in three aspects: Scalability, speedup, and GPU utilization.
The ratio of CPU cores and GPU is 12:1 on single GPU node and 12:2 on multi GPU node. Each process of NAMD occupy one CPU core and 1 GPU. However, GPU can be shared by multiple process. Therefore, it is possible that GPU can be oversubscribed by too many processes and become bottleneck in certain configuration.
B.Performance
Figure 2 shows relative speedup of GPU version compared to CPU only version. Suppose the performance of CPU only version with 1 core is 1x. With 1 CPU core and 1 GPU, NAMD runs 7 times faster than CPU only version. With 2 CPU cores and 1 GPU, NAMD runs 13 times faster. While CPU version scales up well until 12 cores, GPU version hits the peak around 6 cores and starts falling.
 |
| Figure 2 |
Figure 3 shows how utilization of single GPU changes as the number of CPU core increases. With 1 CPU core, GPU uses only 30% of its capability. In other words, 70% of GPU is not doing anything and just stay idle. However, if there are 2 CPU cores, GPU is shared by these 2 cores and overall utilization has been doubled up to around 60%. Likewise, utilization goes over 90% when 4 CPU cores share GPU.
 |
| Figure 3 |
2)Multi GPU environment
Among two graphs in Figure 4, top one shows utilization of first GPU and bottom one shows utilization of second GPU. The fact that two graphs look very similar each other represents NAMD distributes workload evenly to multiple GPU for computation. With 2 CPU cores, utilization of each GPU stays around 40%. With 4 CPU cores, utilization of each GPU goes up over 60%.
 |
| Figure 4 |
GPU application shows different performance charac-teristics from CPU application. Scalability can be affected by the ratio of CPU core and GPU. Oversubscribing GPU can limit the scalability of application. On multi GPU node, the overall speedup is affected by how application distribute workload to each GPU device. At the same time, under-subscribing GPU device, such as workload is not big enough to keep all GPU devices busy, is also limit the performance of GPU application.